- Record: found
- Abstract: found
- Article: found
Multistep diversification in spatiotemporal bacterial-phage coevolution
Read this article at
Abstract
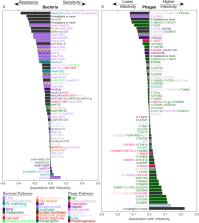
The evolutionary arms race between phages and bacteria, where bacteria evolve resistance to phages and phages retaliate with resistance-countering mutations, is a major driving force of molecular innovation and genetic diversification. Yet attempting to reproduce such ongoing retaliation dynamics in the lab has been challenging; laboratory coevolution experiments of phage and bacteria are typically performed in well-mixed environments and often lead to rapid stagnation with little genetic variability. Here, co-culturing motile E. coli with the lytic bacteriophage T7 on swimming plates, we observe complex spatiotemporal dynamics with multiple genetically diversifying adaptive cycles. Systematically quantifying over 10,000 resistance-infectivity phenotypes between evolved bacteria and phage isolates, we observe diversification into multiple coexisting ecotypes showing a complex interaction network with both host-range expansion and host-switch tradeoffs. Whole-genome sequencing of these evolved phage and bacterial isolates revealed a rich set of adaptive mutations in multiple genetic pathways including in genes not previously linked with phage-bacteria interactions. Synthetically reconstructing these new mutations, we discover phage-general and phage-specific resistance phenotypes as well as a strong synergy with the more classically known phage-resistance mutations. These results highlight the importance of spatial structure and migration for driving phage-bacteria coevolution, providing a concrete system for revealing new molecular mechanisms across diverse phage-bacterial systems.
Abstract
Bacteria and their viruses coexist and coevolve in nature, but maintaining them together in the lab is challenging. Here, a spatially structured environment allowed prolonged coevolution, with bacteria and phage diversifying into multiple ecotypes, uncovering gene mechanisms affecting phage-bacteria interactions.
Related collections
Most cited references81
- Record: found
- Abstract: not found
- Article: not found
Regression Shrinkage and Selection Via the Lasso
- Record: found
- Abstract: found
- Article: not found
Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq.
- Record: found
- Abstract: found
- Article: not found