- Record: found
- Abstract: found
- Article: found
Reassessing the Role of the Type II MqsRA Toxin-Antitoxin System in Stress Response and Biofilm Formation: mqsA Is Transcriptionally Uncoupled from mqsR
Read this article at
Abstract
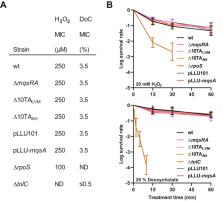
There is growing controversy regarding the role of chromosomal toxin-antitoxin systems in bacterial physiology. mqsRA is a peculiar toxin-antitoxin system, as the gene encoding the toxin precedes that of the antitoxin. This system was previously shown to play a role in stress response and biofilm formation. In this work, we identified two promoters specifically driving the constitutive expression of the antitoxin, thereby decoupling the expression of antitoxin from the toxin. We also showed that mqsRA contributes neither to the regulation of biofilm formation nor to the sensitivity to oxidative stress and bile salts. Finally, we were unable to confirm that the MqsA antitoxin is a global regulator. Altogether, our data are ruling out the involvement of the mqsRA system in Escherichia coli regulatory networks.
ABSTRACT
Toxin-antitoxin (TA) systems are broadly distributed modules whose biological roles remain mostly unknown. The mqsRA system is a noncanonical TA system in which the toxin and antitoxins genes are organized in operon but with the particularity that the toxin gene precedes that of the antitoxin. This system was shown to regulate global processes such as resistance to bile salts, motility, and biofilm formation. In addition, the MqsA antitoxin was shown to be a master regulator that represses the transcription of the csgD, cspD, and rpoS global regulator genes, thereby displaying a pleiotropic regulatory role. Here, we identified two promoters located in the toxin sequence driving the constitutive expression of mqsA, allowing thereby excess production of the MqsA antitoxin compared to the MqsR toxin. Our results show that both antitoxin-specific and operon promoters are not regulated by stresses such as amino acid starvation, oxidative shock, or bile salts. Moreover, we show that the MqsA antitoxin is not a global regulator as suggested, since the expression of csgD, cspD and rpoS is similar in wild-type and Δ mqsRA mutant strains. Moreover, these two strains behave similarly in terms of biofilm formation and sensitivity to oxidative stress or bile salts.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair.
- Record: found
- Abstract: found
- Article: not found
Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli.
- Record: found
- Abstract: found
- Article: not found