- Record: found
- Abstract: found
- Article: found
Exploring the frequency of a TP53 polyadenylation signal variant in tumor DNA from patients diagnosed with lung adenocarcinomas, sarcomas and uterine leiomyomas
Abstract
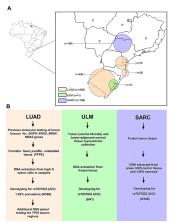
The TP53 3’UTR variant rs78378222 A>C has been detected in different tumor types as a somatic alteration that reduces p53 expression through modification of polyadenylation and miRNA regulation. Its prevalence is not yet known in all tumors. Herein, we examine tumor tissue prevalence of rs7837822 in Brazilian cohorts of patients from south and southeast regions diagnosed with lung adenocarcinoma (LUAD, n=586), sarcoma (SARC, n=188) and uterine leiomyoma (ULM, n=41). The minor allele (C) was identified in heterozygosity in 6/586 LUAD tumors (prevalence = 1.02 %) and none of the SARC and ULM samples. Additionally, next generation sequencing analysis revealed that all variant-positive tumors (n=4) with sample availability had additional pathogenic or likely pathogenic somatic variants in the TP53 coding regions. Among them, 3/4 (75 %) had the same pathogenic or likely pathogenic sequence variant (allele frequency <0.05 in tumor DNA) namely c.751A>C (p.Ile251Leu). Our results indicate a low somatic prevalence of rs78378222 in LUAD, ULM and SARC tumors from Brazilian patients, which suggests that no further analysis of this variant in the specific studied regions of Brazil is warranted. However, these findings should not exclude tumor molecular testing of this TP53 3’UTR functional variant for different populations.
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
Translational control by 5'-untranslated regions of eukaryotic mRNAs.
- Record: found
- Abstract: found
- Article: not found
The role of the 3' untranslated region in post-transcriptional regulation of protein expression in mammalian cells.
- Record: found
- Abstract: found
- Article: not found