- Record: found
- Abstract: found
- Article: found
Integrative epigenomic mapping defines four main chromatin states in Arabidopsis
Read this article at
Abstract
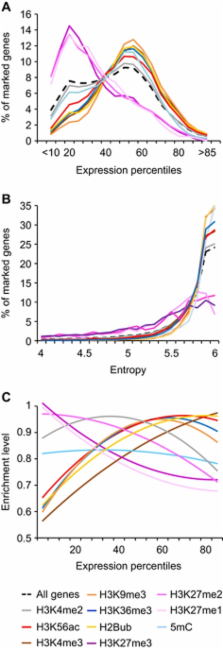
Post-translational modification of histones and DNA methylation are important components of chromatin-level control of genome activity in eukaryotes. However, principles governing the combinatorial association of chromatin marks along the genome remain poorly understood. Here, we have generated epigenomic maps for eight histone modifications (H3K4me2 and 3, H3K27me1 and 2, H3K36me3, H3K56ac, H4K20me1 and H2Bub) in the model plant Arabidopsis and we have combined these maps with others, produced under identical conditions, for H3K9me2, H3K9me3, H3K27me3 and DNA methylation. Integrative analysis indicates that these 12 chromatin marks, which collectively cover ∼90% of the genome, are present at any given position in a very limited number of combinations. Moreover, we show that the distribution of the 12 marks along the genomic sequence defines four main chromatin states, which preferentially index active genes, repressed genes, silent repeat elements and intergenic regions. Given the compact nature of the Arabidopsis genome, these four indexing states typically translate into short chromatin domains interspersed with each other. This first combinatorial view of the Arabidopsis epigenome points to simple principles of organization as in metazoans and provides a framework for further studies of chromatin-based regulatory mechanisms in plants.
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning.
- Record: found
- Abstract: found
- Article: not found
Identification of functional elements and regulatory circuits by Drosophila modENCODE.
- Record: found
- Abstract: found
- Article: not found