- Record: found
- Abstract: found
- Article: found
Roles of gut microbiota in atrial fibrillation: insights from Mendelian randomization analysis and genetic data from over 430,000 cohort study participants
Read this article at
Abstract
Background
Gut microbiota imbalances have been suggested as a contributing factor to atrial fibrillation (AF), but the causal relationship is not fully understood.
Objectives
To explore the causal relationships between the gut microbiota and AF using Mendelian randomization (MR) analysis.
Methods
Summary statistics were from genome-wide association studies (GWAS) of 207 gut microbial taxa (5 phyla, 10 classes, 13 orders, 26 families, 48 genera, and 105 species) (the Dutch Microbiome Project) and two large meta-GWASs of AF. The significant results were validated in FinnGen cohort and over 430,000 UK Biobank participants. Mediation MR analyses were conducted for AF risk factors, including type 2 diabetes, coronary artery disease (CAD), body mass index (BMI), blood lipids, blood pressure, and obstructive sleep apnea, to explore the potential mediation effect of these risk factors in between the gut microbiota and AF.
Results
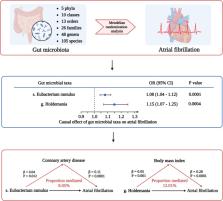
Two microbial taxa causally associated with AF: species Eubacterium ramulus (odds ratio [OR] 1.08, 95% confidence interval [CI] 1.04–1.12, P = 0.0001, false discovery rate (FDR) adjusted p-value = 0.023) and genus Holdemania (OR 1.15, 95% CI 1.07–1.25, P = 0.0004, FDR adjusted p-value = 0.042). Genus Holdemania was associated with incident AF risk in the UK Biobank. The proportion of mediation effect of species Eubacterium ramulus via CAD was 8.05% (95% CI 1.73% − 14.95%, P = 0.008), while the proportion of genus Holdemania on AF via BMI was 12.01% (95% CI 5.17% − 19.39%, P = 0.0005).
Related collections
Most cited references44

- Record: found
- Abstract: found
- Article: found
The UK Biobank resource with deep phenotyping and genomic data
- Record: found
- Abstract: found
- Article: not found