- Record: found
- Abstract: found
- Article: found
Mapping the interactions of the single-stranded DNA binding protein of bacteriophage T4 (gp32) with DNA lattices at single nucleotide resolution: polynucleotide binding and cooperativity
Read this article at
Abstract
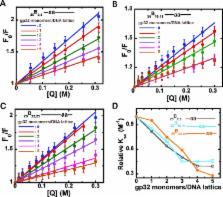
We here use our site-specific base analog mapping approach to study the interactions and binding equilibria of cooperatively-bound clusters of the single-stranded DNA binding protein (gp32) of the T4 DNA replication complex with longer ssDNA (and dsDNA) lattices. We show that in cooperatively bound clusters the binding free energy appears to be equi-partitioned between the gp32 monomers of the cluster, so that all bind to the ssDNA lattice with comparable affinity, but also that the outer domains of the gp32 monomers at the ends of the cluster can fluctuate on and off the lattice and that the clusters of gp32 monomers can slide along the ssDNA. We also show that at very low binding densities gp32 monomers bind to the ssDNA lattice at random, but that cooperatively bound gp32 clusters bind preferentially at the 5′-end of the ssDNA lattice. We use these results and the gp32 monomer-binding results of the companion paper to propose a detailed model for how gp32 might bind to and interact with ssDNA lattices in its various binding modes, and also consider how these clusters might interact with other components of the T4 DNA replication complex.
Related collections
Most cited references26
- Record: found
- Abstract: not found
- Article: not found
Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice.
- Record: found
- Abstract: found
- Article: not found
Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA.
- Record: found
- Abstract: found
- Article: not found