- Record: found
- Abstract: found
- Article: found
Identification of a TuMV isolate (TuMV-ZR) from Pseudostellaria heterophylla and its development into a viral expression vector

Read this article at
Highlights
-
•
A procedure to identify the viral genome was developed with virus-derived sRNAs.
-
•
P. heterophylla TuMV-ZR isolate (TuMV-ZR) were identified.
-
•
TuMV-ZR infectious clone effectively infected the medicinal plant P. heterophylla.
-
•
TuMV-ZR-based vectors continuously expressed foreign genes in P. heterophylla.
Abstract
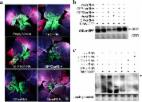
Pseudostellaria heterophylla ( P. heterophylla) is a popular Chinese medicinal herb that is cultivated widely in China. Viral infection is commonly encountered during the production of P. heterophylla. To identify viruses causing P. heterophylla disease, sRNA and mRNA libraries were built for 2 sets of P. heterophylla plants, one set that was planted only once (FGP) and one that was planted three consecutive three times (TGP) in a field, using virus-free tuberous roots as reproductive materials. A comprehensive procedure, including assembling virus-derived sRNA (vsRNA), assessing and cloning the full-length viral genome, building an infectious cloning vector and constructing a virus-based expression vector, was performed to identify viruses infecting P. heterophylla. Ultimately, 48 contig-related viruses were mined from 6 sRNA and 6 mRNA P. heterophylla libraries. A 9762-bp fragment was predicted to be the complete genome of TuMV virus. This sequence was cloned from P. heterophylla, and its infectivity was evaluated using the virus-infection model plant Nicotiana benthamiana ( N. benthamiana) and host plant P. heterophylla. The resulting 9839-bp viral genome was successfully obtained from P. heterophylla and identified as a new P. heterophylla TuMV-ZR isolate. Simultaneously, TuMV-ZR infectious clones were shown to effectively infect P. heterophylla. Furthermore, TuMV-ZR expression vectors were developed, and the ability of a TuMV-ZR-based vector to express foreign genes was determined by analysis with the reporter gene EGFP. TuMV-ZR-based vectors were found to continuously express foreign genes in different organs of P. heterophylla throughout the whole vegetative period. In addition, TuMV-ZR vectors carrying EGFP accumulated in the tuberous roots of P. heterophylla, confirming that tuberous roots are key targets for viral infection and transmission. This study revealed the core pathogenicity of P. heterophylla mosaic virus and developed a new TuMV-ZR-based expression tool that led to long-term protein expression in P. heterophylla, laying the foundation for the identification of the mechanisms of P. heterophylla infection with mosaic viruses and developing tools to express value proteins in the tuberous roots of the medicinal plant P. heterophylla.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
Velvet: algorithms for de novo short read assembly using de Bruijn graphs.
- Record: found
- Abstract: found
- Article: found
RNA virus interference via CRISPR/Cas13a system in plants
- Record: found
- Abstract: found
- Article: not found