- Record: found
- Abstract: found
- Article: found
Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis
Read this article at
Abstract
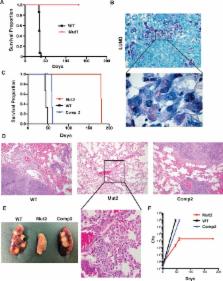
Mycobacterium tuberculosis ( Mtb) mutants lacking rv1411c, which encodes the lipoprotein LprG, and rv1410c, which encodes a putative efflux pump, are dramatically attenuated for growth in mice. Here we show that loss of LprG-Rv1410 in Mtb leads to intracellular triacylglyceride (TAG) accumulation, and overexpression of the locus increases the levels of TAG in the culture medium, demonstrating a role of this locus in TAG transport. LprG binds TAG within a large hydrophobic cleft and is sufficient to transfer TAG from donor to acceptor membranes. Further, LprG-Rv1410 is critical for broadly regulating bacterial growth and metabolism in vitro during carbon restriction and in vivo during infection of mice. The growth defect in mice is due to disrupted bacterial metabolism and occurs independently of key immune regulators. The in vivo essentiality of this locus suggests that this export system and other regulators of metabolism should be considered as targets for novel therapeutics.
Author Summary
Of the estimated 2 billion people worldwide currently infected with Mycobacterium tuberculosis (Mtb), surprisingly few go on to develop active tuberculosis (TB) disease. The vast majority, 95 percent, of infected individuals develop latent TB, remaining infected but without disease. Despite its importance in global health, the question of what determines whether an infected individual will develop active or latent TB remains largely unanswered. Changes in how Mtb grows in response to stressors presented by the host environment likely play an important role in this process. In particular, the manifold ways in which Mtb synthesizes, degrades, and transports lipids dictates its growth in an infected host. Here, we show that lipid transport is an important function of two TB genes known to be required for Mtb’s ability to cause disease in the mouse model of infection. Using a variety of genetic and biochemical techniques, we found that the products of these genes prevent the cytosolic accumulation of a lipid associated with non-growing Mtb under the metabolic conditions it encounters during infection. Our results indicate an important role for the metabolism of Mtb in its ability to orchestrate a productive infection and cause disease.
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
Genetic requirements for mycobacterial survival during infection.
- Record: found
- Abstract: found
- Article: not found
Disseminated tuberculosis in interferon gamma gene-disrupted mice
- Record: found
- Abstract: found
- Article: not found