- Record: found
- Abstract: found
- Article: found
Feedback control of PLK1 by Apolo1 ensures accurate chromosome segregation

Read this article at
SUMMARY
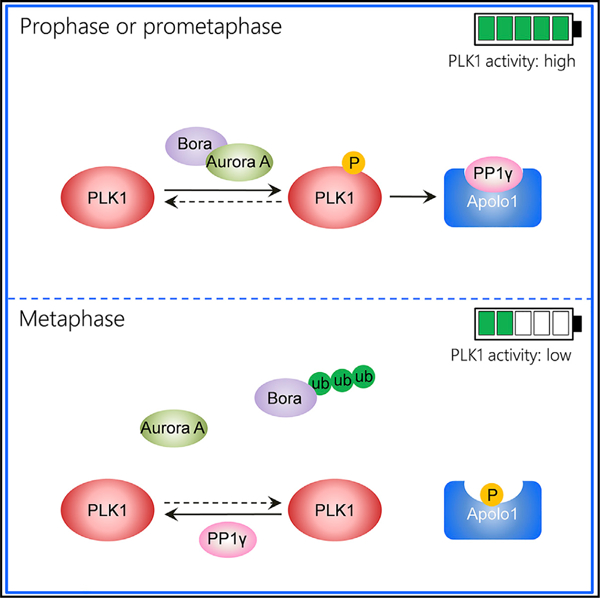
Stable transmission of genetic material during cell division requires accurate chromosome segregation. PLK1 dynamics at kinetochores control establishment of correct kinetochore-microtubule attachments and subsequent silencing of the spindle checkpoint. However, the regulatory mechanism responsible for PLK1 activity in prometaphase has not yet been affirmatively identified. Here we identify Apolo1, which tunes PLK1 activity for accurate kinetochore-microtubule attachments. Apolo1 localizes to kinetochores during early mitosis, and suppression of Apolo1 results in misaligned chromosomes. Using the fluorescence resonance energy transfer (FRET)-based PLK1 activity reporter, we found that Apolo1 sustains PLK1 kinase activity at kinetochores for accurate attachment during prometaphase. Apolo1 is a cognate substrate of PLK1, and the phosphorylation enables PP1γ to inactivate PLK1 by dephosphorylation. Mechanistically, Apolo1 constitutes a bridge between kinase and phosphatase, which governs PLK1 activity in prometaphase. These findings define a previously uncharacterized feedback loop by which Apolo1 provides fine-tuning for PLK1 to guide chromosome segregation in mitosis.
Graphical abstract
In brief
Xu et al. identify Apolo1, which governs PLK1 activity and promotes faithful chromosome segregation in prometaphase by bridging kinase and phosphatase activities.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
A gene-coexpression network for global discovery of conserved genetic modules.
- Record: found
- Abstract: found
- Article: not found
Proteogenomics connects somatic mutations to signaling in breast cancer
- Record: found
- Abstract: found
- Article: not found