- Record: found
- Abstract: found
- Article: found
Gut microbiota functions: metabolism of nutrients and other food components
Read this article at
Abstract
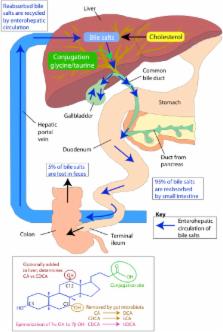
The diverse microbial community that inhabits the human gut has an extensive metabolic repertoire that is distinct from, but complements the activity of mammalian enzymes in the liver and gut mucosa and includes functions essential for host digestion. As such, the gut microbiota is a key factor in shaping the biochemical profile of the diet and, therefore, its impact on host health and disease. The important role that the gut microbiota appears to play in human metabolism and health has stimulated research into the identification of specific microorganisms involved in different processes, and the elucidation of metabolic pathways, particularly those associated with metabolism of dietary components and some host-generated substances. In the first part of the review, we discuss the main gut microorganisms, particularly bacteria, and microbial pathways associated with the metabolism of dietary carbohydrates (to short chain fatty acids and gases), proteins, plant polyphenols, bile acids, and vitamins. The second part of the review focuses on the methodologies, existing and novel, that can be employed to explore gut microbial pathways of metabolism. These include mathematical models, omics techniques, isolated microbes, and enzyme assays.
Related collections
Most cited references168
- Record: found
- Abstract: found
- Article: not found
A core gut microbiome in obese and lean twins
- Record: found
- Abstract: found
- Article: not found
Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease
- Record: found
- Abstract: found
- Article: not found