- Record: found
- Abstract: found
- Article: found
The Core Gut Microbiome of Black Soldier Fly ( Hermetia illucens) Larvae Raised on Low-Bioburden Diets
Read this article at
Abstract
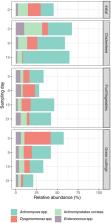
An organism’s gut microbiome handles most of the metabolic processes associated with food intake and digestion but can also strongly affect health and behavior. A stable microbial core community in the gut provides general metabolic competences for substrate degradation and is robust against extrinsic disturbances like changing diets or pathogens. Black Soldier Fly larvae (BSFL; Hermetia illucens) are well known for their ability to efficiently degrade a wide spectrum of organic materials. The ingested substrates build up the high fat and protein content in their bodies that make the larvae interesting for the animal feedstuff industry. In this study, we subjected BSFL to three distinct types of diets carrying a low bioburden and assessed the diets’ impact on larval development and on the composition of the bacterial and archaeal gut community. No significant impact on the gut microbiome across treatments pointed us to the presence of a predominant core community backed by a diverse spectrum of low-abundance taxa. Actinomyces spp., Dysgonomonas spp., and Enterococcus spp. as main members of this community provide various functional and metabolic skills that could be crucial for the thriving of BSFL in various environments. This indicates that the type of diet could play a lesser role in guts of BSFL than previously assumed and that instead a stable autochthonous collection of bacteria provides the tools for degrading of a broad range of substrates. Characterizing the interplay between the core gut microbiome and BSFL helps to understand the involved degradation processes and could contribute to further improving large-scale BSFL rearing.
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample.
- Record: found
- Abstract: found
- Article: not found
The gut bacteria of insects: nonpathogenic interactions.
- Record: found
- Abstract: found
- Article: not found