- Record: found
- Abstract: found
- Article: found
Broadly neutralizing antibodies target a haemagglutinin anchor epitope
Read this article at
Abstract
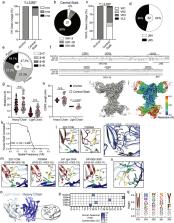
Broadly neutralizing antibodies that target epitopes of haemagglutinin on the influenza virus have the potential to provide near universal protection against influenza virus infection 1 . However, viral mutants that escape broadly neutralizing antibodies have been reported 2, 3 . The identification of broadly neutralizing antibody classes that can neutralize viral escape mutants is critical for universal influenza virus vaccine design. Here we report a distinct class of broadly neutralizing antibodies that target a discrete membrane-proximal anchor epitope of the haemagglutinin stalk domain. Anchor epitope-targeting antibodies are broadly neutralizing across H1 viruses and can cross-react with H2 and H5 viruses that are a pandemic threat. Antibodies that target this anchor epitope utilize a highly restricted repertoire, which encodes two public binding motifs that make extensive contacts with conserved residues in the fusion peptide. Moreover, anchor epitope-targeting B cells are common in the human memory B cell repertoire and were recalled in humans by an oil-in-water adjuvanted chimeric haemagglutinin vaccine 4, 5 , which is a potential universal influenza virus vaccine. To maximize protection against seasonal and pandemic influenza viruses, vaccines should aim to boost this previously untapped source of broadly neutralizing antibodies that are widespread in the human memory B cell pool.
Abstract
A distinct class of broadly neutralizing antibodies to the influenza virus target a membrane-proximal anchor epitope of the haemagglutinin stalk domain.
Related collections
Most cited references96
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.
- Record: found
- Abstract: found
- Article: not found
MUSCLE: multiple sequence alignment with high accuracy and high throughput.
- Record: found
- Abstract: not found
- Article: not found