- Record: found
- Abstract: found
- Article: found
Regulation of transcription factors by sumoylation
Read this article at
ABSTRACT
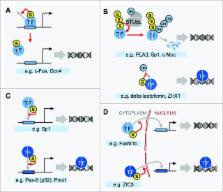
Transcription factors (TFs) are among the most frequently detected targets of sumoylation, and effects of the modification have been studied for about 200 individual TFs to date. TF sumoylation is most often associated with reduced target gene expression, which can be mediated by enhanced interactions with corepressors or by interference with protein modifications that promote transcription. However, recent studies show that sumoylation also regulates gene expression by controlling the levels of TFs that are associated with chromatin. SUMO can mediate this by modulating TF DNA-binding activity, promoting clearance of TFs from chromatin, or indirectly, by influencing TF abundance or localization.
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
PDSM, a motif for phosphorylation-dependent SUMO modification.

- Record: found
- Abstract: found
- Article: found