- Record: found
- Abstract: found
- Article: found
Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean
Read this article at
Abstract
Background
Soybean ( Glycine max [L.] Merr.) is one of the most important oil and protein crops. Ever-increasing soybean consumption necessitates the improvement of varieties for more efficient production. However, both correlations among different traits and genetic interactions among genes that affect a single trait pose a challenge to soybean breeding.
Results
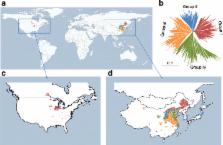
To understand the genetic networks underlying phenotypic correlations, we collected 809 soybean accessions worldwide and phenotyped them for two years at three locations for 84 agronomic traits. Genome-wide association studies identified 245 significant genetic loci, among which 95 genetically interacted with other loci. We determined that 14 oil synthesis-related genes are responsible for fatty acid accumulation in soybean and function in line with an additive model. Network analyses demonstrated that 51 traits could be linked through the linkage disequilibrium of 115 associated loci and these links reflect phenotypic correlations. We revealed that 23 loci, including the known Dt1, E2, E1, Ln, Dt2, Fan, and Fap loci, as well as 16 undefined associated loci, have pleiotropic effects on different traits.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: found
Yield Trends Are Insufficient to Double Global Crop Production by 2050
- Record: found
- Abstract: found
- Article: not found
Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems.
- Record: found
- Abstract: found
- Article: not found