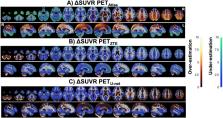
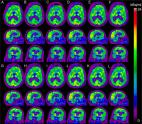
Paul Blanc-Durand:
ORCID: http://orcid.org/0000-0003-1197-2879
Role: ConceptualizationRole: Data curationRole: MethodologyRole: Project administrationRole:
ResourcesRole: ValidationRole: VisualizationRole: Writing – original draftRole: Writing
– review & editing
Maya Khalife: Role: ConceptualizationRole: Data curationRole: MethodologyRole: SupervisionRole:
Writing – review & editing
Brian Sgard: Role: Data curation
Sandeep Kaushik: Role: Formal analysisRole: Software
Marine Soret: Role: Resources
Amal Tiss: Role: Writing – review & editing
Georges El Fakhri: Role: Writing – review & editing
Marie-Odile Habert: Role: MethodologyRole: Writing – review & editing
Florian Wiesinger: Role: Writing – review & editing
Aurélie Kas: Role: ConceptualizationRole: Data curationRole: InvestigationRole: MethodologyRole:
Project administrationRole: SupervisionRole: ValidationRole: Writing – original draftRole:
Writing – review & editing
Stephen D Ginsberg: Role: Editor
Journal ID (nlm-ta): PLoS One
Journal ID (iso-abbrev): PLoS ONE
Journal ID (publisher-id): plos
Journal ID (pmc): plosone
Title:
PLoS ONE
Publisher:
Public Library of Science
(San Francisco, CA USA
)
ISSN
(Electronic):
1932-6203
Publication date Collection: 2019
Publication date
(Electronic):
7
October
2019
Volume: 14
Issue: 10
Electronic Location Identifier: e0223141
Affiliations
[1
]
Nuclear Medicine Department, Groupe Hospitalier Pitié-Salpêtrière C. Foix, APHP, Paris,
France
[2
]
Centre de Neuroimagerie de Recherche (CENIR), Institut du Cerveau et de la Moëlle,
Paris, France
[3
]
GE Healthcare, Bangalore, India
[4
]
Gordon Center for Medical Imaging, Radiology, Massachusetts General Hospital, Harvard
Medical School, Boston, Massachusetts, United States of America
[5
]
Laboratoire d’Imagerie Biomédicale, Sorbonne Université, Paris, France
[6
]
GE Healthcare, Munich, Germany
Nathan S Kline Institute, UNITED STATES
Author notes
Competing Interests: MK received a research grant from GE Healthcare. SK and FW are GE Healthcare employees.
Only non-GE employees had control of inclusion of data and information that might
present a conflict of interest for authors who are employees of GE Healthcare. No
other potential conflict of interest relevant to this article was reported. AK received
honoraria for lectures from GE Healthcare and Piramal. MOH received honoraria for
lectures from Lilly. This does not alter our adherence to PLOS ONE policies on sharing
data and materials. The other authors do not present any conflict of interest.
Author information
Article
Publisher ID:
PONE-D-19-16876
DOI: 10.1371/journal.pone.0223141
PMC ID: 6779234
PubMed ID: 31589623
SO-VID: 9889725e-de41-4352-b6ab-b146e2aa4a7e
Copyright © © 2019 Blanc-Durand et al
License:
This is an open access article distributed under the terms of the
Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided
the original author and source are credited.
The funder provided support in the form of salaries for authors [MK, SK, FW], but
did not have any additional role in the study design, data collection and analysis,
decision to publish. The specific roles of these authors are articulated in the ‘author
contributions’ section.
Subject:
Research Article
Subject:
Medicine and Health Sciences
Subject:
Diagnostic Medicine
Subject:
Diagnostic Radiology
Subject:
Magnetic Resonance Imaging
Subject:
Research and Analysis Methods
Subject:
Imaging Techniques
Subject:
Diagnostic Radiology
Subject:
Magnetic Resonance Imaging
Subject:
Medicine and Health Sciences
Subject:
Radiology and Imaging
Subject:
Diagnostic Radiology
Subject:
Magnetic Resonance Imaging
Subject:
Research and Analysis Methods
Subject:
Imaging Techniques
Subject:
Neuroimaging
Subject:
Positron Emission Tomography
Subject:
Biology and Life Sciences
Subject:
Neuroscience
Subject:
Neuroimaging
Subject:
Positron Emission Tomography
Subject:
Medicine and Health Sciences
Subject:
Diagnostic Medicine
Subject:
Diagnostic Radiology
Subject:
Tomography
Subject:
Positron Emission Tomography
Subject:
Research and Analysis Methods
Subject:
Imaging Techniques
Subject:
Diagnostic Radiology
Subject:
Tomography
Subject:
Positron Emission Tomography
Subject:
Medicine and Health Sciences
Subject:
Radiology and Imaging
Subject:
Diagnostic Radiology
Subject:
Tomography
Subject:
Positron Emission Tomography
Subject:
Research and Analysis Methods
Subject:
Imaging Techniques
Subject:
Neuroimaging
Subject:
Computed Axial Tomography
Subject:
Biology and Life Sciences
Subject:
Neuroscience
Subject:
Neuroimaging
Subject:
Computed Axial Tomography
Subject:
Medicine and Health Sciences
Subject:
Diagnostic Medicine
Subject:
Diagnostic Radiology
Subject:
Tomography
Subject:
Computed Axial Tomography
Subject:
Research and Analysis Methods
Subject:
Imaging Techniques
Subject:
Diagnostic Radiology
Subject:
Tomography
Subject:
Computed Axial Tomography
Subject:
Medicine and Health Sciences
Subject:
Radiology and Imaging
Subject:
Diagnostic Radiology
Subject:
Tomography
Subject:
Computed Axial Tomography
Subject:
Research and Analysis Methods
Subject:
Imaging Techniques
Subject:
Neuroimaging
Subject:
Biology and Life Sciences
Subject:
Neuroscience
Subject:
Neuroimaging
Subject:
Research and Analysis Methods
Subject:
Imaging Techniques
Subject:
Biology and Life Sciences
Subject:
Anatomy
Subject:
Brain
Subject:
Cerebral Cortex
Subject:
Cerebellum
Subject:
Medicine and Health Sciences
Subject:
Anatomy
Subject:
Brain
Subject:
Cerebral Cortex
Subject:
Cerebellum
Subject:
Computer and Information Sciences
Subject:
Neural Networks
Subject:
Biology and Life Sciences
Subject:
Neuroscience
Subject:
Neural Networks
Subject:
Biology and Life Sciences
Subject:
Anatomy
Subject:
Musculoskeletal System
Subject:
Skeleton
Subject:
Skull
Subject:
Mastoid Process
Subject:
Medicine and Health Sciences
Subject:
Anatomy
Subject:
Musculoskeletal System
Subject:
Skeleton
Subject:
Skull
Subject:
Mastoid Process