- Record: found
- Abstract: found
- Article: found
Plasmid-mediated fluoroquinolone resistance associated with extra-intestinal Escherichia coli isolates from hospital samples
Read this article at
Abstract
Background & objectives:
Infection from fluoroquinolone-resistant extra-intestinal Escherichia coli is a global concern. In this study, isolation and characterization of fluoroquinolone-resistant extra-intestinal E. coli isolates obtained from hospital samples were undertaken to detect plasmid-mediated quinolone resistance ( PMQR) genes.
Methods:
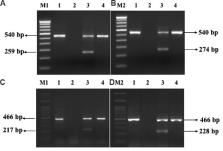
Forty three isolates of E. coli obtained from patients with extra-intestinal infections were subjected to antibiogram to detect fluoroquinolone resistance. The mechanism of fluoroquinolone resistance was determined by the detection of PMQR genes and mutations in quinolone resistance determining region (QRDR).
Results:
Of the 43 isolates, 36 were resistant to nalidixic acid (83.72%) and 28 to ciprofloxacin (65.11%). Eight E. coli isolates showed total resistance to both the antimicrobials without any minimum inhibitory concentration. The detection of PMQR genes with qnr primers showed the presence of qnrA in two, qnrB in six and qnrS in 21 isolates. The gene coding for quinolone efflux pump ( qepA) was not detected in any of the isolates tested. The presence of some unexpressed PMQR genes in fluoroquinolone sensitive isolates was also observed.
Interpretation & conclusions:
The detection of silent PMQR genes as observed in the present study presents a risk of the transfer of the silent resistance genes to other microorganisms if present in conjugative plasmids, thus posing a therapeutic challenge to the physicians. Hence, frequent monitoring is to be done for all resistance determinants.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
The worldwide emergence of plasmid-mediated quinolone resistance.

- Record: found
- Abstract: found
- Article: found
Food Reservoir for Escherichia coli Causing Urinary Tract Infections
- Record: found
- Abstract: found
- Article: not found