- Record: found
- Abstract: found
- Article: found
Relationship Between Odor Intensity Estimates and COVID-19 Prevalence Prediction in a Swedish Population
Read this article at
Abstract
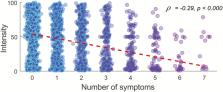
In response to the coronavirus disease 2019 (COVID-19) pandemic, countries have implemented various strategies to reduce and slow the spread of the disease in the general population. For countries that have implemented restrictions on its population in a stepwise manner, monitoring of COVID-19 prevalence is of importance to guide the decision on when to impose new, or when to abolish old, restrictions. We are here determining whether measures of odor intensity in a large sample can serve as one such measure. Online measures of how intense common household odors are perceived and symptoms of COVID-19 were collected from 2440 Swedes. Average odor intensity ratings were then compared to predicted COVID-19 population prevalence over time in the Swedish population and were found to closely track each other ( r = −0.83). Moreover, we found that there was a large difference in rated intensity between individuals with and without COVID-19 symptoms and the number of symptoms was related to odor intensity ratings. Finally, we found that individuals progressing from reporting no symptoms to subsequently reporting COVID-19 symptoms demonstrated a large drop in olfactory performance. These data suggest that measures of odor intensity, if obtained in a large and representative sample, can be used as an indicator of COVID-19 disease in the general population. Importantly, this simple measure could easily be implemented in countries without widespread access to COVID-19 testing or implemented as a fast early response before widespread testing can be facilitated.
Related collections
Most cited references19
- Record: found
- Abstract: found
- Article: not found
A Novel Coronavirus from Patients with Pneumonia in China, 2019

- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found