- Record: found
- Abstract: found
- Article: found
ER as master regulator of membrane trafficking and organelle function
Read this article at
Abstract
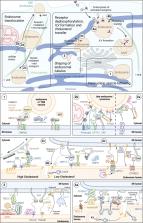
Wenzel et al. review how the endoplasmic reticulum controls the positioning, dynamics, and functions of other organelles via membrane contact sites.
Abstract
The endoplasmic reticulum (ER), which occupies a large portion of the cytoplasm, is the cell’s main site for the biosynthesis of lipids and carbohydrate conjugates, and it is essential for folding, assembly, and biosynthetic transport of secreted proteins and integral membrane proteins. The discovery of abundant membrane contact sites (MCSs) between the ER and other membrane compartments has revealed that, in addition to its biosynthetic and secretory functions, the ER plays key roles in the regulation of organelle dynamics and functions. In this review, we will discuss how the ER regulates endosomes, lysosomes, autophagosomes, mitochondria, peroxisomes, and the Golgi apparatus via MCSs. Such regulation occurs via lipid and Ca 2+ transfer and also via control of in trans dephosphorylation reactions and organelle motility, positioning, fusion, and fission. The diverse controls of other organelles via MCSs manifest the ER as master regulator of organelle biology.
Related collections
Most cited references204
- Record: found
- Abstract: found
- Article: not found
Autophagy: renovation of cells and tissues.

- Record: found
- Abstract: found
- Article: found
Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum
- Record: found
- Abstract: found
- Article: not found