- Record: found
- Abstract: found
- Article: found
ICTV Virus Taxonomy Profile: Herpesviridae 2021
Read this article at
Abstract
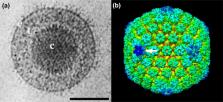
Members of the family Herpesviridae have enveloped, spherical virions with characteristic complex structures consisting of symmetrical and non-symmetrical components. The linear, double-stranded DNA genomes of 125–241 kbp contain 70–170 genes, of which 43 have been inherited from an ancestral herpesvirus. In general, herpesviruses have coevolved with and are highly adapted to their hosts, which comprise many mammalian, avian and reptilian species. Following primary infection, they are able to establish lifelong latent infection, during which there is limited viral gene expression. Severe disease is usually observed only in the foetus, the very young, the immunocompromised or following infection of an alternative host. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Herpesviridae, which is available at ictv.global/report/herpesviridae.
Related collections
Most cited references5
- Record: found
- Abstract: found
- Article: not found
Three-dimensional structure of herpes simplex virus from cryo-electron tomography.
- Record: found
- Abstract: found
- Article: not found
Topics in herpesvirus genomics and evolution.

- Record: found
- Abstract: found
- Article: found