- Record: found
- Abstract: found
- Article: found
Genetic diversities and phylogenetic analyses of three Chinese main ethnic groups in southwest China: A Y-Chromosomal STR study
Read this article at
Abstract
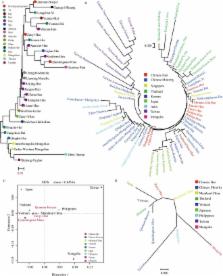
Short tandem repeats (STRs) located on the Y chromosome with the properties of male-specific inheritance and haploidy are widely used in forensics to analyze paternal genealogies and match male trace donors to evidence. Besides, Y-chromosomal haplotypes play an important role in providing breathtaking insights into population genetic history. However, the genetic diversity and forensic characteristics of Y-STRs in Guizhou main ethnic groups (Hans, Miaos and Bouyeis) remain uncharacterized. Here, we obtained Y-chromosomal 23-marker haplotypes in three Guizhou populations and submitted the first batch of Y-STR haplotype data to the YHRD. The HD in the aforementioned three populations are 0.99990, 0.99983, and 0.99979, respectively, and DC values are 0.9902, 0.9908, and 0.97959, respectively. Subsequently, genetic differentiation between our newly studied populations and reference groups along ethnic/administrative divisions, as well as national/continental boundaries were investigated via AMOVA, MDS, and phylogenetic relationship reconstruction. Significant genetic differentiations from our subjects and other groups are identified in ethnically, linguistically and geographically diverse populations, including most prominently Tibetans and Uyghurs among 30 mainland Chinese populations, Taiwanese groups and others among 58 Asian populations, as well as African groups and others among 89 worldwide populations. Qiannan Bouyei has a close genetic relationship with Guangxi Zhuang, and Zunyi Han and Qiandongnan Miao have close genetic affinity with Hunan Han and Guizhou Shui, respectively. Collectively, this new-generation Y-STR amplification system can be used as a supplementary tool in forensic identification and male parentage testing and even pedigree search.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
Tracing the peopling of the world through genomics
- Record: found
- Abstract: found
- Article: not found
Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences.
- Record: found
- Abstract: found
- Article: not found