- Record: found
- Abstract: found
- Article: found
Sulindac selectively induces autophagic apoptosis of GABAergic neurons and alters motor behaviour in zebrafish
Read this article at
Abstract
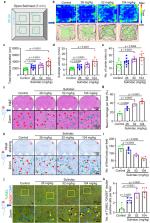
Nonsteroidal anti-inflammatory drugs compose one of the most widely used classes of medications, but the risks for early development remain controversial, especially in the nervous system. Here, we utilized zebrafish larvae to assess the potentially toxic effects of nonsteroidal anti-inflammatory drugs and found that sulindac can selectively induce apoptosis of GABAergic neurons in the brains of zebrafish larvae brains. Zebrafish larvae exhibit hyperactive behaviour after sulindac exposure. We also found that akt1 is selectively expressed in GABAergic neurons and that SC97 (an Akt1 activator) and exogenous akt1 mRNA can reverse the apoptosis caused by sulindac. Further studies showed that sulindac binds to retinoid X receptor alpha (RXRα) and induces autophagy in GABAergic neurons, leading to activation of the mitochondrial apoptotic pathway. Finally, we verified that sulindac can lead to hyperactivity and selectively induce GABAergic neuron apoptosis in mice. These findings suggest that excessive use of sulindac may lead to early neurodevelopmental toxicity and increase the risk of hyperactivity, which could be associated with damage to GABAergic neurons.
Abstract
Nonsteroidal anti-inflammatory drugs (NSAIDs) are widely used but their risks for early neurodevelopment remain controversial. Here, the authors showed in zebrafish larvae that sulindac induces GABAergic neuron apoptosis through autophagy activation that leads to hyperactive behavior.
Related collections
Most cited references97
- Record: found
- Abstract: found
- Article: not found
Integrating single-cell transcriptomic data across different conditions, technologies, and species
- Record: found
- Abstract: found
- Article: not found
Fast, sensitive, and accurate integration of single cell data with Harmony
- Record: found
- Abstract: found
- Article: not found