- Record: found
- Abstract: found
- Article: found
Metabolic engineering of Saccharomyces cerevisiae to produce 1-hexadecanol from xylose
Read this article at
Abstract
Background
An advantageous but challenging approach to overcome the limited supply of petroleum and relieve the greenhouse effect is to produce bulk chemicals from renewable materials. Fatty alcohols, with a billion-dollar global market, are important raw chemicals for detergents, emulsifiers, lubricants, and cosmetics production. Microbial production of fatty alcohols has been successfully achieved in several industrial microorganisms. However, most of the achievements were using glucose, an edible sugar, as the carbon source. To produce fatty alcohols in a renewable manner, non-edible sugars such as xylose will be a more appropriate feedstock.
Results
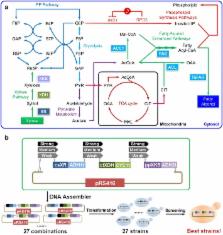
In this study, we aim to engineer a Saccharomyces cerevisiae strain that can efficiently convert xylose to fatty alcohols. To this end, we first introduced the fungal xylose utilization pathway consisting of xylose reductase (XR), xylitol dehydrogenase (XDH), and xylulose kinase (XKS) into a fatty alcohol-producing S. cerevisiae strain (XF3) that was developed in our previous studies to achieve 1-hexadecanol production from xylose at 0.4 g/L. We next applied promoter engineering on the xylose utilization pathway to optimize the expression levels of XR, XDH, and XKS, and increased the 1-hexadecanol titer by 171 %. To further improve the xylose-based fatty alcohol production, two optimized S. cerevisiae strains from promoter engineering were evolved with the xylose as the sole carbon source. We found that the cell growth rate was improved at the expense of decreased fatty alcohol production, which indicated 1-hexadecanol was mainly produced as a non-growth associated product. Finally, through fed-batch fermentation, we successfully achieved 1-hexadecanol production at over 1.2 g/L using xylose as the sole carbon source, which represents the highest titer of xylose-based 1-hexadecanol reported in microbes to date.
Conclusions
A fatty alcohol-producing S. cerevisiae strain was engineered in this study to produce 1-hexadecanol from xylose. Although the xylose pathway we developed in this study could be further improved, this proof-of-concept study, for the first time to our best knowledge, demonstrated that the xylose-based fatty alcohol could be produced in S. cerevisiae with potential applications in developing consolidated bioprocessing for producing other fatty acid-derived chemicals.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels.

- Record: found
- Abstract: found
- Article: found
DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways

- Record: found
- Abstract: found
- Article: found