- Record: found
- Abstract: found
- Article: found
Molecular networks of the FOXP2 transcription factor in the brain
review-article

Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
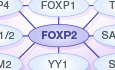
The discovery of the FOXP2 transcription factor, and its implication in a rare severe human speech and language disorder, has led to two decades of empirical studies focused on uncovering its roles in the brain using a range of in vitro and in vivo methods. Here, we discuss what we have learned about the regulation of FOXP2, its downstream effectors, and its modes of action as a transcription factor in brain development and function, providing an integrated overview of what is currently known about the critical molecular networks.
Abstract
Related collections
Most cited references176
- Record: found
- Abstract: found
- Article: not found
Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder
Ditte Demontis, Raymond K Walters, Joanna Martin … (2019)
- Record: found
- Abstract: found
- Article: not found
Foxp3 programs the development and function of CD4+CD25+ regulatory T cells.
- Record: found
- Abstract: found
- Article: not found
Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations
Brian O'Roak, Pelagia Deriziotis, Choli Lee … (2011)