- Record: found
- Abstract: found
- Article: found
Identification and Characterization of mRNA and lncRNA Expression Profiles in Age-Related Hearing Loss
Read this article at
Abstract
Objectives
Age-related hearing loss (ARHL), or presbycusis, is caused by disorders of sensory hair cells and auditory neurons. Many studies have suggested that the accumulation of mitochondrial DNA damage, the production of reactive oxygen species, noise, inflammation, and decreased antioxidant function are associated with subsequent cochlear senescence in response to aging stress. Long non-coding RNA (lncRNA) has been reported to play important roles in various diseases. However, the function of lncRNA in ARHL remains unclear. In this study, we analyzed the common expression profiles of messenger RNA (mRNA) and lncRNA through ARHL-related RNA-sequencing datasets.
Methods
We selected and downloaded three different sets of RNA-sequencing data for ARHL. We performed differential expression analysis to find common mRNA and lncRNA profiles in the cochleae of aged mice compared to young mice. Gene Ontology (GO) analysis was used for functional exploration. Real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed to validate mRNAs and lncRNAs. In addition, we performed trans target prediction analysis with differentially expressed mRNAs and lncRNAs to understand the function of these mRNAs and lncRNAs in ARHL.
Results
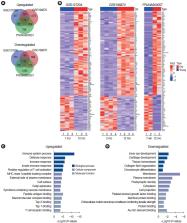
We identified 112 common mRNAs and 10 common lncRNAs in the cochleae of aged mice compared to young mice. GO analysis showed that the 112 upregulated mRNAs were enriched in the defense response pathway. When we performed qRT-PCR with 1 mM H 2O 2-treated House Ear Institute-Organ of Corti 1 (HEI-OC1) cells, the qRT-PCR results were consistent with the RNA-sequencing analysis data. lncRNA-mRNA networks were constructed using the 10 common lncRNAs and 112 common mRNAs in ARHL.
Conclusion
Our study provides a comprehensive understanding of the common mRNA and lncRNA expression profiles in ARHL. Knowledge of ARHL-associated mRNAs and lncRNAs could be useful for better understanding ARHL and these mRNAs and lncRNAs might be a potential therapeutic target for preventing ARHL.
Related collections
Most cited references34

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found