- Record: found
- Abstract: found
- Article: found
Quantifying differences in fMRI preprocessing pipelines via OGRE (One-step General Registration and Extraction)
Read this article at
Abstract
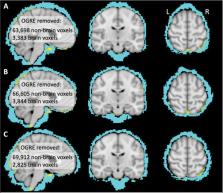
Volumetric preprocessing methods continue to enjoy great popularity in the analysis of functional MRI (fMRI) data. Among these methods, the software packages FSL (FMRIB, Oxford, UK) and FreeSurfer (LCN, Charlestown, MA) are omnipresent throughout the field. However, it remains unknown what advantages an integrated FSL+FreeSurfer preprocessing approach might provide over FSL alone. Here we developed the One-step General Registration and Extraction (OGRE) pipeline to combine FreeSurfer and FSL tools for brain extraction and registration, for FSL volumetric analysis of fMRI data. We compared preprocessing approaches in a dataset wherein adult human volunteers (N=26) performed a precision drawing task during fMRI scanning. OGRE’s preprocessing, compared to traditional FSL preprocessing, led to lower inter-individual variability across the brain, more precise brain extraction, and greater detected activation in sensorimotor areas contralateral to movement. This demonstrates that the introduction of FreeSurfer tools via OGRE preprocessing can improve fMRI data analysis, in the context of FSL’s volumetric analysis approach. The OGRE pipeline provides a turnkey method to integrate FreeSurfer-based brain extraction and registration with FSL analysis of task fMRI data.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
The organization of the human cerebral cortex estimated by intrinsic functional connectivity.
- Record: found
- Abstract: found
- Article: not found
Advances in functional and structural MR image analysis and implementation as FSL.
- Record: found
- Abstract: found
- Article: not found