- Record: found
- Abstract: found
- Article: found
Role of HPV 16 variants among cervical carcinoma samples from Northeastern Brazil
Read this article at
Abstract
Background
Cervical cancer is the fourth most common type of cancer affecting women globally. In Brazil, it is the third most frequent type of cancer in women and HPV is present in approximately 90% of cases. Evidence suggests that variants of HPV 16 can interfere biologically and etiologically during the development of cervical cancer.
Methods
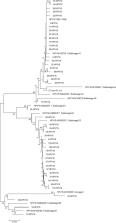
Cervix tumor fragments were collected, their DNA was extracted, and nested PCR was used to detect HPV. Positive samples were sequenced to determine the viral genotype. To characterize the HPV 16 strains, positive samples PCR was used to amplify the LCR and E6 regions of the HPV 16 virus.
Results
Data from 120 patients with cervical cancer were analyzed. Most women were between 41 and 54 years of age, had schooling until primary school, a family income between 1 and 2 times the minimum wage and were married/in a consensual union. There was no statistically significant association between HPV or socio-demographic variables and risk factors for cervical cancer ( P < 0.05). HPV was present in 88 women (73%). The most prevalent types were HPV 16 (53.4%), HPV 18 (13.8%), HPV 35 (6.9%) and HPV 45 (5.7%). Of the 47 HPV 16 positive cases, variant A (49%) was present in 23 samples, followed by variant D in 20 cases (43%), and variants B and C in 2 cases each (4%). The most prevalent histological type of HPV 16 tumors was squamous cell carcinoma, followed by adenocarcinoma. There was a statistically significant association between HPV 16 variants and the tumors’ histological types ( P < 0.001).
Related collections
Most cited references25

- Record: found
- Abstract: found
- Article: found
Human papillomavirus molecular biology and disease association
- Record: found
- Abstract: found
- Article: not found
Human papillomavirus genome variants.
- Record: found
- Abstract: found
- Article: not found