- Record: found
- Abstract: found
- Article: found
Thymic Epithelial Cell Alterations and Defective Thymopoiesis Lead to Central and Peripheral Tolerance Perturbation in MHCII Deficiency
Read this article at
Abstract
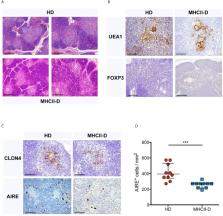
Major Histocompatibility Complex (MHC) class II (MHCII) deficiency (MHCII-D), also known as Bare Lymphocyte Syndrome (BLS), is a rare combined immunodeficiency due to mutations in genes regulating expression of MHCII molecules. MHCII deficiency results in impaired cellular and humoral immune responses, leading to severe infections and autoimmunity. Abnormal cross-talk with developing T cells due to the absence of MHCII expression likely leads to defects in thymic epithelial cells (TEC). However, the contribution of TEC alterations to the pathogenesis of this primary immunodeficiency has not been well characterized to date, in particular in regard to immune dysregulation. To this aim, we have performed an in-depth cellular and molecular characterization of TEC in this disease. We observed an overall perturbation of thymic structure and function in both MHCII −/− mice and patients. Transcriptomic and proteomic profiling of murine TEC revealed several alterations. In particular, we demonstrated that impairment of lymphostromal cross-talk in the thymus of MHCII −/− mice affects mTEC maturation and promiscuous gene expression and causes defects of central tolerance. Furthermore, we observed peripheral tolerance impairment, likely due to defective Treg cell generation and/or function and B cell tolerance breakdown. Overall, our findings reveal disease-specific TEC defects resulting in perturbation of central tolerance and limiting the potential benefits of hematopoietic stem cell transplantation in MHCII deficiency.
Related collections
Most cited references59

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Near-optimal probabilistic RNA-seq quantification.
- Record: found
- Abstract: found
- Article: not found