- Record: found
- Abstract: found
- Article: found
MAIT cell activation and dynamics associated with COVID-19 disease severity
Read this article at
Abstract
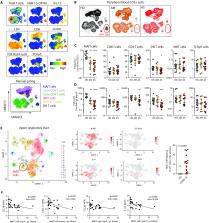
MAIT cell activation and decline in blood are associated with COVID-19 severity, features that dynamically recover in convalescence.
Innate-like rapid responders
Viral infections elicit host responses from conventional T cells, innate lymphoid cells, and innate-like lymphocyte subsets. Parrot et al. used blood from acute and convalescent COVID-19 patients to investigate how SARS-CoV-2 infection affects the innate-like mucosa-associated invariant T (MAIT) cells. Acute viral infection induced a profound decline in the number of blood MAIT cells and activation of the residual blood MAIT cells. The loss of circulating MAIT cells in acute COVID-19 patients coincided with enrichment of MAIT cells among T cells recovered from the respiratory tract. With convalescence, the number of blood MAIT cells and their activation status reverted toward normal. These findings indicate that circulating MAIT cells are mobilized early after SARS-CoV-2 infection and may contribute to both resolution and exacerbation of COVID-19–associated pneumonia.
Abstract
Severe coronavirus disease 2019 (COVID-19) is characterized by excessive inflammation of the lower airways. The balance of protective versus pathological immune responses in COVID-19 is incompletely understood. Mucosa-associated invariant T (MAIT) cells are antimicrobial T cells that recognize bacterial metabolites and can also function as innate-like sensors and mediators of antiviral responses. Here, we investigated the MAIT cell compartment in COVID-19 patients with moderate and severe disease, as well as in convalescence. We show profound and preferential decline in MAIT cells in the circulation of patients with active disease paired with strong activation. Furthermore, transcriptomic analyses indicated substantial MAIT cell enrichment and proinflammatory IL-17A bias in the airways. Unsupervised analysis identified MAIT cell CD69 high and CXCR3 low immunotypes associated with poor clinical outcome. MAIT cell levels normalized in the convalescent phase, consistent with dynamic recruitment to the tissues and later release back into the circulation when disease is resolved. These findings indicate that MAIT cells are engaged in the immune response against SARS-CoV-2 and suggest their possible involvement in COVID-19 immunopathogenesis.
Related collections
Most cited references54
- Record: found
- Abstract: found
- Article: found
The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3).
- Record: found
- Abstract: found
- Article: found
Remdesivir for the Treatment of Covid-19 — Final Report
- Record: found
- Abstract: found
- Article: not found