- Record: found
- Abstract: found
- Article: not found
Encephalitis and Death in Wild Mammals at a Rehabilitation Center after Infection with Highly Pathogenic Avian Influenza A(H5N8) Virus, United Kingdom
Read this article at
Abstract
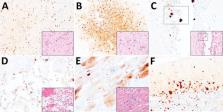
We report a disease and mortality event involving swans, seals, and a fox at a wildlife rehabilitation center in the United Kingdom during late 2020. Five swans had onset of highly pathogenic avian influenza virus infection while in captivity. Subsequently, 5 seals and a fox died (or were euthanized) after onset of clinical disease. Avian-origin influenza A virus subtype H5N8 was retrospectively determined as the cause of disease. Infection in the seals manifested as seizures, and immunohistochemical and molecular testing on postmortem samples detected a neurologic distribution of viral products. The fox died overnight after sudden onset of inappetence, and postmortem tissues revealed neurologic and respiratory distribution of viral products. Live virus was isolated from the swans, seals, and the fox, and a single genetic change was detected as a potential adaptive mutation in the mammalian-derived viral sequences. No human influenza-like illness was reported in the weeks after the event.
Related collections
Most cited references30

- Record: found
- Abstract: found
- Article: found
Fast and accurate short read alignment with Burrows–Wheeler transform
- Record: found
- Abstract: found
- Article: found
Twelve years of SAMtools and BCFtools
- Record: found
- Abstract: found
- Article: not found
