- Record: found
- Abstract: found
- Article: not found
Precise therapeutic gene correction by a simple nuclease-induced double-strand break
Read this article at
Abstract
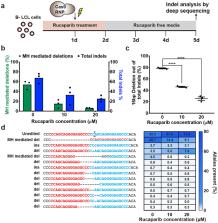
Current programmable nuclease-based (e.g. CRISPR-Cas9) methods for precise correction of a disease-causing genetic mutation harness the Homology Directed Repair (HDR) pathway. However, this repair process requires co-delivery of an exogenous DNA donor to recode the sequence and can be inefficient in many cell types. Here, we show that disease-causing frameshift mutations resulting from microduplications can be efficiently reverted to the wild-type sequence simply by generating a double-strand break (DSB) near the center of the duplication. We demonstrate this in patient-derived cell lines for two diseases: Limb-Girdle Muscular Dystrophy 2G (LGMD2G) 1 and Hermansky-Pudlak Syndrome Type 1 (HPS1) 2 . Clonal analysis of Streptococcus pyogenes Cas9 (SpyCas9) nuclease-treated LGMD2G iPSCs revealed that ~80% contained at least one wild-type allele and that this correction restored TCAP expression in LGMD2G iPSC-derived myotubes. Efficient genotypic correction was also observed upon SpyCas9 treatment of an HPS1 patient-derived B-lymphoblastoid cell line (B-LCL). Inhibition of PARP-1 (poly (ADP-ribose) polymerase) suppresses the nuclease-mediated collapse of the microduplication to the wild-type sequence, confirming that precise correction is mediated by the MMEJ (microhomology-mediated end joining) pathway. Analysis of editing by SpyCas9 and Lachnospiraceae bacterium ND2006 Cas12a (LbaCas12a) at non-pathogenic microduplications within the genome that range in length from 4 bp to 36 bp indicates that the correction strategy is broadly applicable to a wide range of microduplication lengths and can be initiated by a variety of nucleases. The simplicity, reliability and efficacy of this MMEJ-based therapeutic strategy should permit the development of nuclease-based gene correction therapies for a variety of diseases that are associated with microduplications.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements
- Record: found
- Abstract: found
- Article: not found
