- Record: found
- Abstract: found
- Article: not found
A METTL3-METTL14 complex mediates mammalian nuclear RNA N 6-adenosine methylation
research-article
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
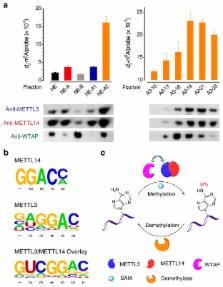
N 6-methyladenosine (m 6A) is the most prevalent and reversible internal modification in mammalian messenger and non-coding RNAs. We report here that human METTL14 catalyzes m 6A RNA methylation. Together with METTL3, the only previously known m 6A methyltransferase, these two proteins form a stable heterodimer core complex of METTL3-14 that functions in cellular m 6A deposition on mammalian nuclear RNAs. WTAP, a mammalian splicing factor, can interact with this complex and affect this methylation.
Related collections
Most cited references11
- Record: found
- Abstract: found
- Article: not found
MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor.
Silin Zhong, Hongying Li, Zsuzsanna Bodi … (2008)
- Record: found
- Abstract: found
- Article: not found
Reversible RNA adenosine methylation in biological regulation.
Guifang Jia, Ye Fu, Tong-Chuan He (2013)
- Record: found
- Abstract: found
- Article: not found
Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene.
Mary J. Clancy, Mary Shambaugh, Candace Timpte … (2002)
