- Record: found
- Abstract: found
- Article: found
Implementing the CRISPR/Cas9 Technology in Eucalyptus Hairy Roots Using Wood-Related Genes
Read this article at
Abstract
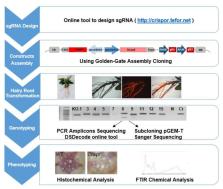
Eucalypts are the most planted hardwoods worldwide. The availability of the Eucalyptus grandis genome highlighted many genes awaiting functional characterization, lagging behind because of the lack of efficient genetic transformation protocols. In order to efficiently generate knock-out mutants to study the function of eucalypts genes, we implemented the powerful CRISPR/Cas9 gene editing technology with the hairy roots transformation system. As proofs-of-concept, we targeted two wood-related genes: Cinnamoyl-CoA Reductase1 ( CCR1), a key lignin biosynthetic gene and IAA9A an auxin dependent transcription factor of Aux/IAA family. Almost all transgenic hairy roots were edited but the allele-editing rates and spectra varied greatly depending on the gene targeted. Most edition events generated truncated proteins, the prevalent edition types were small deletions but large deletions were also quite frequent. By using a combination of FT-IR spectroscopy and multivariate analysis (partial least square analysis (PLS-DA)), we showed that the CCR1-edited lines, which were clearly separated from the controls. The most discriminant wave-numbers were attributed to lignin. Histochemical analyses further confirmed the decreased lignification and the presence of collapsed vessels in CCR1-edited lines, which are characteristics of CCR1 deficiency. Although the efficiency of editing could be improved, the method described here is already a powerful tool to functionally characterize eucalypts genes for both basic research and industry purposes.
Related collections
Most cited references54

- Record: found
- Abstract: found
- Article: found
mixOmics: An R package for ‘omics feature selection and multiple data integration
- Record: found
- Abstract: found
- Article: found
CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture
- Record: found
- Abstract: not found
- Article: not found