- Record: found
- Abstract: found
- Article: found
Molecular typing of Cyclospora cayetanensis in produce and clinical samples using targeted enrichment of complete mitochondrial genomes and next-generation sequencing
Read this article at
Abstract
Background
Outbreaks of cyclosporiasis, a diarrheal illness caused by Cyclospora cayetanensis, have been a public health issue in the USA since the mid 1990’s. In 2018, 2299 domestically acquired cases of cyclosporiasis were reported in the USA as a result of multiple large outbreaks linked to different fresh produce commodities. Outbreak investigations are hindered by the absence of standardized molecular epidemiological tools for C. cayetanensis. For other apicomplexan coccidian parasites, multicopy organellar DNA such as mitochondrial genomes have been used for detection and molecular typing.
Methods
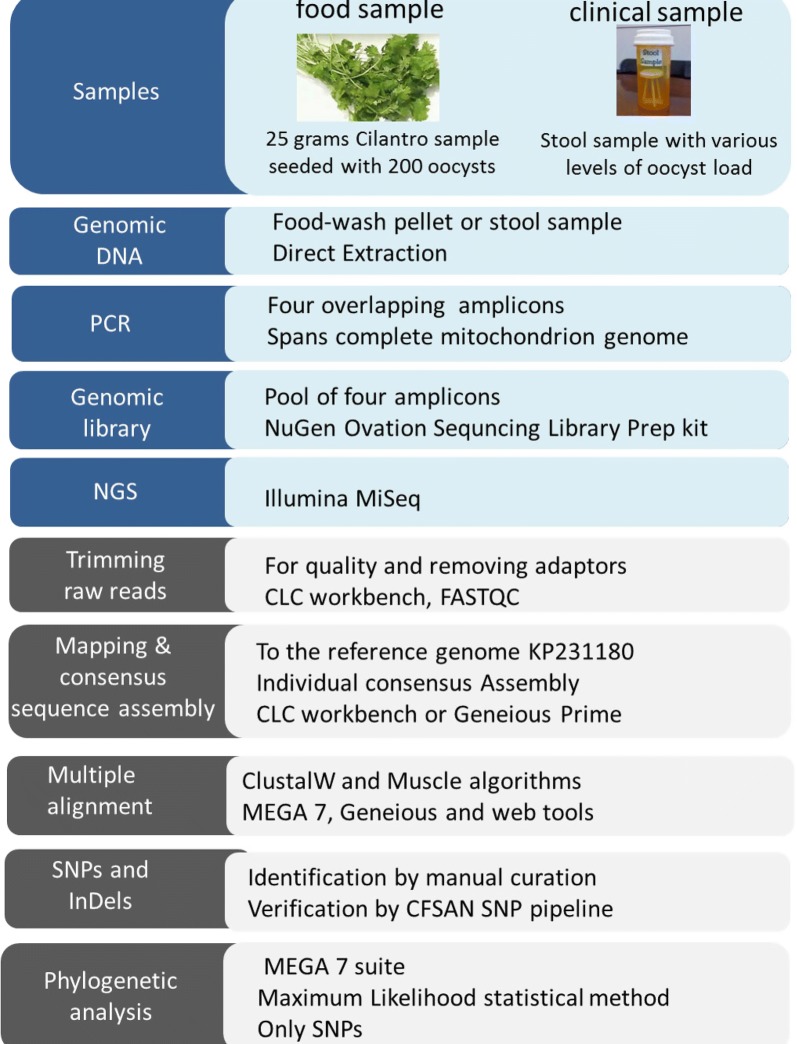
We developed a workflow to obtain complete mitochondrial genome sequences from cilantro samples and clinical samples for typing of C. cayetanensis isolates. The 6.3 kb long C. cayetanensis mitochondrial genome was amplified by PCR in four overlapping amplicons from genomic DNA extracted from cilantro, seeded with oocysts, and from stool samples positive for C. cayetanensis by diagnostic methods. DNA sequence libraries of pooled amplicons were prepared and sequenced via next-generation sequencing (NGS). Sequence reads were assembled using a custom bioinformatics pipeline.
Results
This approach allowed us to sequence complete mitochondrial genomes from the samples studied. Sequence alterations, such as single nucleotide polymorphism (SNP) profiles and insertion and deletions (InDels), in mitochondrial genomes of 24 stool samples from patients with cyclosporiasis diagnosed in 2014, exhibited discriminatory power. The cluster dendrogram that was created based on distance matrices of the complete mitochondrial genome sequences, indicated distinct strain-level diversity among the 2014 C. cayetanensis outbreak isolates analyzed in this study.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Mutation pressure and the evolution of organelle genomic architecture.
- Record: found
- Abstract: found
- Article: not found
Practical Value of Food Pathogen Traceability through Building a Whole-Genome Sequencing Network and Database.
- Record: found
- Abstract: found
- Article: not found