- Record: found
- Abstract: found
- Article: found
Chemically programmable bacterial probes for the recognition of cell surface proteins
Read this article at
Abstract
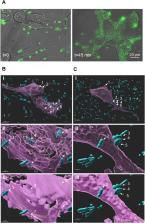
Common methods to label cell surface proteins (CSPs) involve the use of fluorescently modified antibodies (Abs) or small-molecule-based ligands. However, optimizing the labeling efficiency of such systems, for example, by modifying them with additional fluorophores or recognition elements, is challenging. Herein we show that effective labeling of CSPs overexpressed in cancer cells and tissues can be obtained with fluorescent probes based on chemically modified bacteria. The bacterial probes (B-probes) are generated by non-covalently linking a bacterial membrane protein to DNA duplexes appended with fluorophores and small-molecule binders of CSPs overexpressed in cancer cells. We show that B-probes are exceptionally simple to prepare and modify because they are generated from self-assembled and easily synthesized components, such as self-replicating bacterial scaffolds and DNA constructs that can be readily appended, at well-defined positions, with various types of dyes and CSP binders. This structural programmability enabled us to create B-probes that can label different types of cancer cells with distinct colors, as well as generate very bright B-probes in which the multiple dyes are spatially separated along the DNA scaffold to avoid self-quenching. This enhancement in the emission signal enabled us to label the cancer cells with greater sensitivity and follow the internalization of the B-probes into these cells. The potential to apply the design principles underlying B-probes in therapy or inhibitor screening is also discussed here.
Graphical abstract
Decorating bacteria with modified DNA duplexes is presented as a general strategy to obtain fluorescent bacterial probes (B-probes) that can selectively identify cancer cells. The high structural programmability of such systems enabled the creation of B-probes that can selectively label cancer cells and tissues with different emission colors and with enhanced brightness.
Related collections
Most cited references76
- Record: found
- Abstract: found
- Article: not found
The fluorescent toolbox for assessing protein location and function.

- Record: found
- Abstract: found
- Article: found
Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy

- Record: found
- Abstract: found
- Article: found
