- Record: found
- Abstract: found
- Article: found
Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
An algorithm for progressive multiple alignment of sequences with insertions.
Ari Löytynoja, Nick Goldman (2005)
- Record: found
- Abstract: found
- Article: not found
Prediction of lipoprotein signal peptides in Gram-negative bacteria.
- Record: found
- Abstract: found
- Article: found
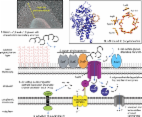
Complex Glycan Catabolism by the Human Gut Microbiota: The Bacteroidetes Sus-like Paradigm*
Eric C. Martens, Nicole M. Koropatkin, Thomas Smith … (2009)