- Record: found
- Abstract: found
- Article: found
mRNA transcript quantification in archival samples using multiplexed, color-coded probes
Read this article at
Abstract
Background
A recently developed probe-based technology, the NanoString nCounter™ gene expression system, has been shown to allow accurate mRNA transcript quantification using low amounts of total RNA. We assessed the ability of this technology for mRNA expression quantification in archived formalin-fixed, paraffin-embedded (FFPE) oral carcinoma samples.
Results
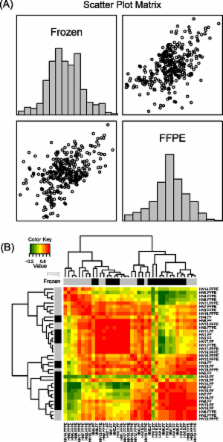
We measured the mRNA transcript abundance of 20 genes ( COL3A1, COL4A1, COL5A1, COL5A2, CTHRC1, CXCL1, CXCL13, MMP1, P4HA2, PDPN, PLOD2, POSTN, SDHA, SERPINE1, SERPINE2, SERPINH1, THBS2, TNC, GAPDH, RPS18) in 38 samples (19 paired fresh-frozen and FFPE oral carcinoma tissues, archived from 1997-2008) by both NanoString and SYBR Green I fluorescent dye-based quantitative real-time PCR (RQ-PCR). We compared gene expression data obtained by NanoString vs. RQ-PCR in both fresh-frozen and FFPE samples. Fresh-frozen samples showed a good overall Pearson correlation of 0.78, and FFPE samples showed a lower overall correlation coefficient of 0.59, which is likely due to sample quality. We found a higher correlation coefficient between fresh-frozen and FFPE samples analyzed by NanoString (r = 0.90) compared to fresh-frozen and FFPE samples analyzed by RQ-PCR (r = 0.50). In addition, NanoString data showed a higher mean correlation (r = 0.94) between individual fresh-frozen and FFPE sample pairs compared to RQ-PCR (r = 0.53).
Conclusions
Based on our results, we conclude that both technologies are useful for gene expression quantification in fresh-frozen or FFPE tissues; however, the probe-based NanoString method achieved superior gene expression quantification results when compared to RQ-PCR in archived FFPE samples. We believe that this newly developed technique is optimal for large-scale validation studies using total RNA isolated from archived, FFPE samples.
Related collections
Most cited references12
- Record: found
- Abstract: found
- Article: not found
Direct multiplexed measurement of gene expression with color-coded probe pairs.
- Record: found
- Abstract: not found
- Article: not found
Thirteen Ways to Look at the Correlation Coefficient
- Record: found
- Abstract: found
- Article: not found