- Record: found
- Abstract: found
- Article: found
Role of the Multidrug Resistance Efflux Pump MexCD-OprJ in the Pseudomonas aeruginosa Quorum Sensing Response
Read this article at
Abstract
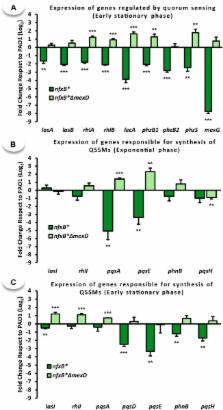
Multidrug efflux pumps constitute a category of antibiotic resistance determinants that are a part of the core bacterial genomes. Given their conservation, it is conceivable that they present functions beyond the extrusion of antibiotics currently used for therapy. Pseudomonas aeruginosa stands as a relevant respiratory pathogen, with a high prevalence at hospitals and in cystic fibrosis patients. Part of its success relies on its low susceptibility to antibiotics and on the production of virulence factors, whose expression is regulated in several cases by quorum sensing (QS). We found that overexpression of the MexCD-OprJ multidrug efflux pump shuts down the P. aeruginosa QS response. Our results support that MexCD-OprJ extrudes kynurenine, a precursor of the alkyl-quinolone signal (AQS) molecules. Anthranilate and octanoate, also AQS precursors, do not seem to be extruded by MexCD-OprJ. Kynurenine extrusion is not sufficient to reduce the QS response in a mutant overexpressing this efflux pump. Impaired QS response is mainly due to the extrusion of 4-hydroxy-2-heptylquinoline (HHQ), the precursor of the Pseudomonas Quinolone Signal (PQS), leading to low PQS intracellular levels and reduced production of QS signal molecules. As the consequence, the expression of QS-regulated genes is impaired and the production of QS-regulated virulence factors strongly decreases in a MexCD-OprN P. aeruginosa overexpressing mutant. Previous work showed that MexEF-OprJ, another P. aeruginosa efflux pump, is also able of extruding kynurenine and HHQ. However, opposite to our findings, the QS defect in a MexEF-OprN overproducer is due to kynurenine extrusion. These results indicate that, although efflux pumps can share some substrates, the affinity for each of them can be different. Although the QS response is triggered by population density, information on additional elements able of modulating such response is still scarce. This is particularly important in the case of P. aeruginosa lung chronic infections, a situation in which QS-defective mutants are accumulated. If MexCD-OprJ overexpression alleviates the cost associated to triggering the QS response when un-needed, it could be possible that MexCD-OprJ antibiotic resistant overproducer strains might be selected even in the absence of antibiotic selective pressure, acting as antibiotic resistant cheaters in heterogeneous P. aeruginosa populations.
Related collections
Most cited references91
- Record: found
- Abstract: found
- Article: not found
Multidrug-resistance efflux pumps - not just for resistance.
- Record: found
- Abstract: found
- Article: not found