- Record: found
- Abstract: found
- Article: not found
Protein Expression Profile of ACE2 in the Normal and COVID-19-Affected Human Brain
Read this article at
Abstract
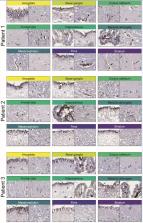
SARS-coronavirus 2 (SARS-CoV-2) that caused the coronavirus disease 2019 (COVID-19) pandemic has posed to be a global challenge. An increasing number of neurological symptoms have been linked to the COVID-19 disease, but the underlying mechanisms of such symptoms and which patients could be at risk are not yet established. The suggested key receptor for host cell entry is angiotensin I converting enzyme 2 (ACE2). Previous studies on limited tissue material have shown no or low protein expression of ACE2 in the normal brain. Here, we used stringently validated antibodies and immunohistochemistry to examine the protein expression of ACE2 in all major regions of the normal brain. The expression pattern was compared with the COVID-19-affected brain of patients with a varying degree of neurological symptoms. In the normal brain, the expression was restricted to the choroid plexus and ependymal cells with no expression in any other brain cell types. Interestingly, in the COVID-19-affected brain, an upregulation of ACE2 was observed in endothelial cells of certain patients, most prominently in the white matter and with the highest expression observed in the patient with the most severe neurological symptoms. The data shows differential expression of ACE2 in the diseased brain and highlights the need to further study the role of endothelial cells in COVID-19 disease in relation to neurological symptoms.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: found
COVID-19: consider cytokine storm syndromes and immunosuppression
- Record: found
- Abstract: found
- Article: not found
Proteomics. Tissue-based map of the human proteome.

- Record: found
- Abstract: found
- Article: found
