- Record: found
- Abstract: found
- Article: not found
Investigation of structural analogs of hydroxychloroquine for SARS-CoV-2 main protease (Mpro): A computational drug discovery study
Read this article at
Abstract
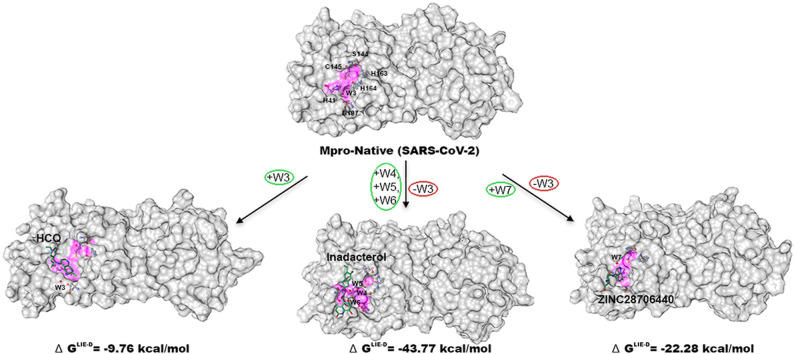
The main protease (Mpro) is the key enzyme of nCOVID-19 and plays a decisive role that makes it an attractive drug target. Multiple analysis of crystal structures reveal the presence of W1, W2, and W3 water locations in the active site pocket of Mpro; W1 and W2 are unstable and are weakly bonded with protein in comparison to W3 of Mpro-native. So, we adopt the water displacement method to occupy W1 or W2 sites by triggering HCQ or its analogs to inactivate the enzyme. Virtual screening is employed to find out best analogs of HCQ, molecular docking is used for water displacement from catalytic region of Mpro, and finally, MD simulations are conducted for validation of these findings. The docking study reveals that W1 and W2 are occupied by respective atoms of ZINC28706440 whereas W2 by HCQ and indacaterol. Finally, MD results demonstrate (i) HCQ occupies W1 and W2 positions but its analogs (indacaterol and ZINC28706440) are inadequate to retain either W1 or W2 (ii) His41 and Asp187 are stabilized by W3 in Mpro-native and His41, Cys145 and HCQ by W7 in ZINC28706440, and W4, W5, and W6 make water mediated bridge between indacaterol with His41. The structural, dynamical, and thermodynamic (WFP and J value) profiling parameters suggest that W3, W4, and W7 are prominent in their corresponding positions in comparison with W5 and W6. The final results conclude that ZINC28706440 may act as a best analog of HCQ with acceptable physico-chemical and toxicological scores and may further be synthesized for experimental validation.
Graphical abstract
Related collections
Most cited references62
- Record: found
- Abstract: not found
- Article: not found
VMD: Visual molecular dynamics
- Record: found
- Abstract: found
- Article: not found
AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading.
- Record: found
- Abstract: found
- Article: not found