- Record: found
- Abstract: found
- Article: found
Mechanisms of Functional Pleiotropy of p73 in Cancer and Beyond
Read this article at
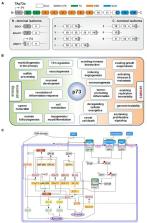
Abstract
The transcription factor p73 is a structural and functional homolog of TP53, the most famous and frequently mutated tumor-suppressor gene. The TP73 gene can synthesize an overwhelming number of isoforms via splicing events in 5′ and 3′ ends and alternative promoter usage. Although it originally came into the spotlight due to the potential of several of these isoforms to mimic p53 functions, it is now clear that TP73 has its own unique identity as a master regulator of multifaceted processes in embryonic development, tissue homeostasis, and cancer. This remarkable functional pleiotropy is supported by a high degree of mechanistic heterogeneity, which extends far-beyond the typical mode of action by transactivation and largely relies on the ability of p73 isoforms to form protein–protein interactions (PPIs) with a variety of nuclear and cytoplasmic proteins. Importantly, each p73 isoform carries a unique combination of functional domains and residues that facilitates the establishment of PPIs in a highly selective manner. Herein, we summarize the expanding functional repertoire of TP73 in physiological and oncogenic processes. We emphasize how TP73’s ability to control neurodevelopment and neurodifferentiation is co-opted in cancer cells toward neoneurogenesis, an emerging cancer hallmark, whereby tumors promote their own innervation. By further exploring the canonical and non-canonical mechanistic patterns of p73, we apprehend its functional diversity as the result of a sophisticated and coordinated interplay of: (a) the type of p73 isoforms (b) the presence of p73 interaction partners in the cell milieu, and (c) the architecture of target gene promoters. We suppose that dysregulation of one or more of these parameters in tumors may lead to cancer initiation and progression by reactivating p73 isoforms and/or p73-regulated differentiation programs thereof in a spatiotemporally inappropriate manner. A thorough understanding of the mechanisms supporting p73 functional diversity is of paramount importance for the efficient and precise p73 targeting not only in cancer, but also in other pathological conditions where TP73 dysregulation is causally involved.
Related collections
Most cited references110
- Record: found
- Abstract: found
- Article: not found
Comprehensive genomic profiles of small cell lung cancer.
- Record: found
- Abstract: found
- Article: not found
Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma.
- Record: found
- Abstract: found
- Article: not found