- Record: found
- Abstract: found
- Article: not found
An Interferon-Inducible Neutrophil-Driven Blood Transcriptional Signature in Human Tuberculosis
Read this article at
Abstract
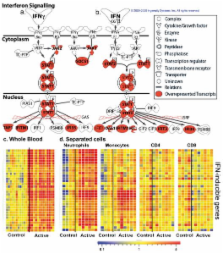
Tuberculosis (TB), caused by infection with Mycobacterium tuberculosis ( M. tuberculosis), is a major cause of morbidity and mortality worldwide and efforts to control TB are hampered by difficulties with diagnosis, prevention and treatment 1, 2. Most people infected with M. tuberculosis remain asymptomatic, termed latent TB, with a 10% lifetime risk of developing active TB disease, but current tests cannot identify which individuals will develop disease 3. The immune response to M. tuberculosis is complex and incompletely characterized, hindering development of new diagnostics, therapies and vaccines 4, 5. We identified a whole blood 393 transcript signature for active TB in intermediate and high burden settings, correlating with radiological extent of disease and reverting to that of healthy controls following treatment. A subset of latent TB patients had signatures similar to those in active TB patients. We also identified a specific 86-transcript signature that discriminated active TB from other inflammatory and infectious diseases. Modular and pathway analysis revealed that the TB signature was dominated by a neutrophil-driven interferon (IFN)-inducible gene profile, consisting of both IFN-γ and Type I IFNαβ signalling. Comparison with transcriptional signatures in purified cells and flow cytometric analysis, suggest that this TB signature reflects both changes in cellular composition and altered gene expression. Although an IFN signature was also observed in whole blood of patients with Systemic Lupus Erythematosus (SLE), their complete modular signature differed from TB with increased abundance of plasma cell transcripts. Our studies demonstrate a hitherto under-appreciated role of Type I IFNαβ signalling in TB pathogenesis, which has implications for vaccine and therapeutic development. Our study also provides a broad range of transcriptional biomarkers with potential as diagnostic and prognostic tools to combat the TB epidemic.
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus.
- Record: found
- Abstract: found
- Article: not found
Genetic dissection of immunity to mycobacteria: the human model.
- Record: found
- Abstract: found
- Article: not found
