- Record: found
- Abstract: found
- Article: found
Effect of mutations on binding of ligands to guanine riboswitch probed by free energy perturbation and molecular dynamics simulations
Read this article at
Abstract
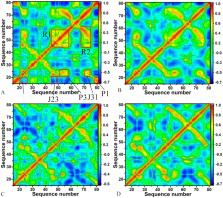
Riboswitches can regulate gene expression by direct and specific interactions with ligands and have recently attracted interest as potential drug targets for antibacterial. In this work, molecular dynamics (MD) simulations, free energy perturbation (FEP) and molecular mechanics generalized Born surface area (MM-GBSA) methods were integrated to probe the effect of mutations on the binding of ligands to guanine riboswitch (GR). The results not only show that binding free energies predicted by FEP and MM-GBSA obtain an excellent correlation, but also indicate that mutations involved in the current study can strengthen the binding affinity of ligands GR. Residue-based free energy decomposition was applied to compute ligand-nucleotide interactions and the results suggest that mutations highly affect interactions of ligands with key nucleotides U22, U51 and C74. Dynamics analyses based on MD trajectories indicate that mutations not only regulate the structural flexibility but also change the internal motion modes of GR, especially for the structures J12, J23 and J31, which implies that the aptamer domain activity of GR is extremely plastic and thus readily tunable by nucleotide mutations. This study is expected to provide useful molecular basis and dynamics information for the understanding of the function of GR and possibility as potential drug targets for antibacterial.
Related collections
Most cited references74
- Record: found
- Abstract: found
- Article: not found
MMPBSA.py: An Efficient Program for End-State Free Energy Calculations.
- Record: found
- Abstract: not found
- Article: not found