- Record: found
- Abstract: found
- Article: found
Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
Read this article at
Abstract
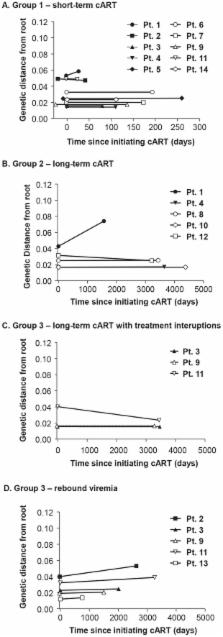
A better understanding of changes in HIV-1 population genetics with combination antiretroviral therapy (cART) is critical for designing eradication strategies. We therefore analyzed HIV-1 genetic variation and divergence in patients' plasma before cART, during suppression on cART, and after viral rebound. Single-genome sequences of plasma HIV-1 RNA were obtained from HIV-1 infected patients prior to cART (N = 14), during suppression on cART (N = 14) and/or after viral rebound following interruption of cART (N = 5). Intra-patient population diversity was measured by average pairwise difference (APD). Population structure was assessed by phylogenetic analyses and a test for panmixia. Measurements of intra-population diversity revealed no significant loss of overall genetic variation in patients treated for up to 15 years with cART. A test for panmixia, however, showed significant changes in population structure in 2/10 patients after short-term cART (<1 year) and in 7/10 patients after long-term cART (1–15 years). The changes consisted of diverse sets of viral variants prior to cART shifting to populations containing one or more genetically uniform subpopulations during cART. Despite these significant changes in population structure, rebound virus after long-term cART had little divergence from pretherapy virus, implicating long-lived cells infected before cART as the source for rebound virus. The appearance of genetically uniform virus populations and the lack of divergence after prolonged cART and cART interruption provide strong evidence that HIV-1 persists in long-lived cells infected before cART was initiated, that some of these infected cells may be capable of proliferation, and that on-going cycles of viral replication are not evident.
Author Summary
Anti-HIV compounds are highly effective for preventing the onset of AIDS but they do not cure infected individuals. Very low levels of virus remain detectable in the blood of most patients despite antiviral treatment and levels surge if treatment is stopped. It is crucial to understand why current treatments are not equipped to cure HIV infection so that new therapies addressing these shortcomings can be developed. By characterizing genetic sequences of HIV in patients before and during antiviral treatment, we found that the low levels of virus detected in the blood of treated patients did not result from newly infected cells but originated from cells, or the daughters of cells, that were already infected when treatment was initiated. This finding demonstrates that HIV present in blood after prolonged antiviral treatment is derived from cells infected prior to treatment which likely expanded over time through cell division. Such long lived, infected cells are likely the critical target for developing strategies to cure HIV infection.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy.
- Record: found
- Abstract: found
- Article: not found
Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy.
- Record: found
- Abstract: found
- Article: not found