- Record: found
- Abstract: found
- Article: found
Genomes of cultivated and wild Capsicum species provide insights into pepper domestication and population differentiation
Read this article at
Abstract
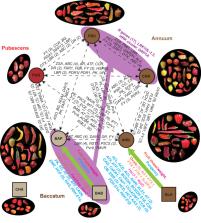
Pepper ( Capsicum spp.) is one of the earliest cultivated crops and includes five domesticated species, C. annuum var. annuum, C. chinense, C. frutescens, C. baccatum var. pendulum and C. pubescens. Here, we report a pepper graph pan-genome and a genome variation map of 500 accessions from the five domesticated Capsicum species and close wild relatives. We identify highly differentiated genomic regions among the domesticated peppers that underlie their natural variations in flowering time, characteristic flavors, and unique resistances to biotic and abiotic stresses. Domestication sweeps detected in C. annuum var. annuum and C. baccatum var. pendulum are mostly different, and the common domestication traits, including fruit size, shape and pungency, are achieved mainly through the selection of distinct genomic regions between these two cultivated species. Introgressions from C. baccatum into C. chinense and C. frutescens are detected, including those providing genetic sources for various biotic and abiotic stress tolerances.
Abstract
Existing genetics and genomics studies of peppers mainly focus on single species. Here, the authors report a pepper graph pan-genome and a genome variation map of 500 accessions from five domesticated species and close wild relatives to reveal their domestication, introgression and population differentiation.
Related collections
Most cited references89

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found
IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies
- Record: found
- Abstract: found
- Article: not found