- Record: found
- Abstract: found
- Article: found
A SUMO-Dependent Protein Network Regulates Chromosome Congression during Oocyte Meiosis
Read this article at
Summary
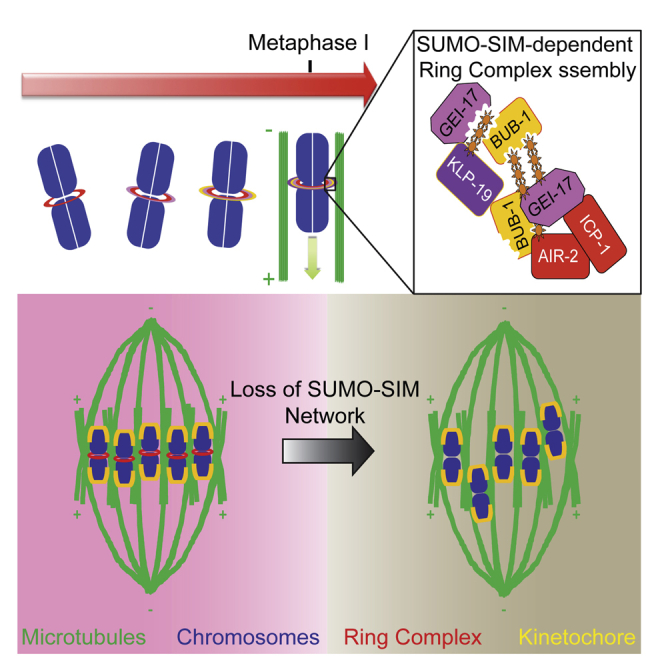
During Caenorhabditis elegans oocyte meiosis, a multi-protein ring complex (RC) localized between homologous chromosomes, promotes chromosome congression through the action of the chromokinesin KLP-19. While some RC components are known, the mechanism of RC assembly has remained obscure. We show that SUMO E3 ligase GEI-17/PIAS is required for KLP-19 recruitment to the RC, and proteomic analysis identified KLP-19 as a SUMO substrate in vivo. In vitro analysis revealed that KLP-19 is efficiently sumoylated in a GEI-17-dependent manner, while GEI-17 undergoes extensive auto-sumoylation. GEI-17 and another RC component, the kinase BUB-1, contain functional SUMO interaction motifs (SIMs), allowing them to recruit SUMO modified proteins, including KLP-19, into the RC. Thus, dynamic SUMO modification and the presence of SIMs in RC components generate a SUMO-SIM network that facilitates assembly of the RC. Our results highlight the importance of SUMO-SIM networks in regulating the assembly of dynamic protein complexes.
Graphical Abstract
Highlights
-
•
SUMO conjugates accumulate in the ring complex within the midbivalent during meiosis
-
•
Ring complex assembly requires active sumoylation
-
•
The SUMO E3 ligase GEI-17/PIAS sumoylates the chromokinesin KLP-19 in vivo and in vitro
-
•
Meiotic chromosome congression depends on sumoylation and non-covalent SUMO binding
Abstract
Pelisch et al. provide evidence that highly dynamic, coordinated, and spatially constrained sumoylation regulates chromosome congression during meiosis in C. elegans oocytes.
Related collections
Most cited references38

- Record: found
- Abstract: found
- Article: found
The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans
- Record: found
- Abstract: found
- Article: not found
Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair.

- Record: found
- Abstract: found
- Article: found