- Record: found
- Abstract: found
- Article: found
Network target for screening synergistic drug combinations with application to traditional Chinese medicine
Read this article at
Abstract
Background
Multicomponent therapeutics offer bright prospects for the control of complex diseases in a synergistic manner. However, finding ways to screen the synergistic combinations from numerous pharmacological agents is still an ongoing challenge.
Results
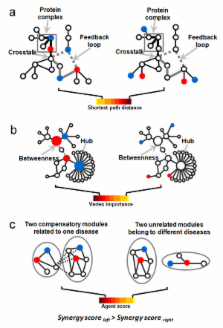
In this work, we proposed for the first time a “network target”-based paradigm instead of the traditional "single target"-based paradigm for virtual screening and established an algorithm termed NIMS (Network target-based Identification of Multicomponent Synergy) to prioritize synergistic agent combinations in a high throughput way. NIMS treats a disease-specific biological network as a therapeutic target and assumes that the relationship among agents can be transferred to network interactions among the molecular level entities (targets or responsive gene products) of agents. Then, two parameters in NIMS, Topology Score and Agent Score, are created to evaluate the synergistic relationship between each given agent combinations. Taking the empirical multicomponent system traditional Chinese medicine (TCM) as an illustrative case, we applied NIMS to prioritize synergistic agent pairs from 63 agents on a pathological process instanced by angiogenesis. The NIMS outputs can not only recover five known synergistic agent pairs, but also obtain experimental verification for synergistic candidates combined with, for example, a herbal ingredient Sinomenine, which outperforms the meet/min method. The robustness of NIMS was also showed regarding the background networks, agent genes and topological parameters, respectively. Finally, we characterized the potential mechanisms of multicomponent synergy from a network target perspective.
Conclusions
NIMS is a first-step computational approach towards identification of synergistic drug combinations at the molecular level. The network target-based approaches may adjust current virtual screen mode and provide a systematic paradigm for facilitating the development of multicomponent therapeutics as well as the modernization of TCM.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: found
Statistical mechanics of complex networks
- Record: found
- Abstract: found
- Article: not found