- Record: found
- Abstract: found
- Article: not found
Innate and Adaptive Immunity through Autophagy
Read this article at
Abstract
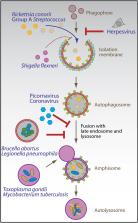
The two main proteolytic machineries of eukaryotic cells, lysosomes and proteasomes, receive substrates by different routes. Polyubiquitination targets proteins for proteasomal degradation, whereas autophagy delivers intracellular material for lysosomal hydrolysis. The importance of autophagy for cell survival has long been appreciated, but more recently, its essential role in both innate and adaptive immunity has been characterized. Autophagy is now recognized to restrict viral infections and replication of intracellular bacteria and parasites. Additionally, this pathway delivers cytoplasmic antigens for MHC class II presentation to the adaptive immune system, which then in turn is able to regulate autophagy. At the same time, autophagy plays a role in the survival and the cell death of T cells. Thus, the immune system utilizes autophagic degradation of cytoplasmic material, to both restrict intracellular pathogens and regulate adaptive immunity.
Related collections
Most cited references77
- Record: found
- Abstract: found
- Article: not found
A ubiquitin-like system mediates protein lipidation.
- Record: found
- Abstract: found
- Article: not found
SYFPEITHI: database for MHC ligands and peptide motifs.
- Record: found
- Abstract: not found
- Article: not found
