- Record: found
- Abstract: found
- Article: found
Bioinformatic Analyses of Unique (Orphan) Core Genes of the Genus Acidithiobacillus: Functional Inferences and Use As Molecular Probes for Genomic and Metagenomic/Transcriptomic Interrogation
Read this article at
Abstract
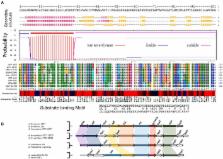
Using phylogenomic and gene compositional analyses, five highly conserved gene families have been detected in the core genome of the phylogenetically coherent genus Acidithiobacillus of the class Acidithiobacillia. These core gene families are absent in the closest extant genus Thermithiobacillus tepidarius that subtends the Acidithiobacillus genus and roots the deepest in this class. The predicted proteins encoded by these core gene families are not detected by a BLAST search in the NCBI non-redundant database of more than 90 million proteins using a relaxed cut-off of 1.0e −5. None of the five families has a clear functional prediction. However, bioinformatic scrutiny, using pI prediction, motif/domain searches, cellular location predictions, genomic context analyses, and chromosome topology studies together with previously published transcriptomic and proteomic data, suggests that some may have functions associated with membrane remodeling during cell division perhaps in response to pH stress. Despite the high level of amino acid sequence conservation within each family, there is sufficient nucleotide variation of the respective genes to permit the use of the DNA sequences to distinguish different species of Acidithiobacillus, making them useful additions to the armamentarium of tools for phylogenetic analysis. Since the protein families are unique to the Acidithiobacillus genus, they can also be leveraged as probes to detect the genus in environmental metagenomes and metatranscriptomes, including industrial biomining operations, and acid mine drainage (AMD).
Related collections
Most cited references73
- Record: found
- Abstract: found
- Article: not found
MRBAYES: Bayesian inference of phylogenetic trees.
- Record: found
- Abstract: found
- Article: not found