- Record: found
- Abstract: found
- Article: found
Low-cesium rice: mutation in OsSOS2 reduces radiocesium in rice grains
Read this article at
Abstract
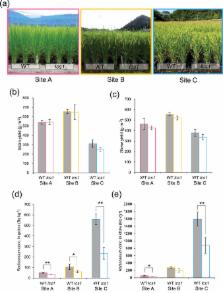
In Japan, radiocesium contamination in foods has become of great concern and it is a primary issue to reduce grain radiocesium concentration in rice ( Oryza sativa L.). Here, we report a low-cesium rice mutant 1 ( lcs1) with the radiocesium concentration in grain about half that in the wild-type cultivar. Genetic analyses revealed that a mutation in OsSOS2, which encodes a serine/threonine-protein kinase required for the salt overly sensitive (SOS) pathway in plants, is responsible for the decreased cesium (Cs) concentrations in lcs1. Physiological analyses showed that Cs + uptake by lcs1 roots was significantly decreased under low-potassium (K +) conditions in the presence of sodium (Na +) (low K +/Na +). The transcript levels of several K + and Na + transporter genes, such as OsHAK1, OsHAK5, OsAKT1, and OsHKT2;1 were significantly down-regulated in lcs1 grown at low K +/Na +. The decreased Cs + uptake in lcs1 might be closely related to the lower expression of these genes due to the K +/Na + imbalance in the lcs1 roots caused by the OsSOS2 mutation. Since the lcs1 plant had no significant negative effects on agronomic traits when grown in radiocesium-contaminated paddy fields, this mutant could be used directly in agriculture for reducing radiocesium in rice grains.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: found
A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data
- Record: found
- Abstract: found
- Article: not found
The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter.
- Record: found
- Abstract: found
- Article: not found