- Record: found
- Abstract: found
- Article: found
Multiple types of programmed necrosis such as necroptosis, pyroptosis, oxytosis/ferroptosis, and parthanatos contribute simultaneously to retinal damage after ischemia–reperfusion
Read this article at
Abstract
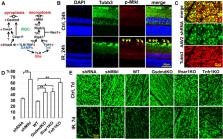
Ischemia–reperfusion (IR) injury is implicated in a large array of pathological conditions in the retina. Increasing experimental evidence suggests that programmed necrosis makes a significant contribution to inflammation and retinal damage triggered by IR. Since there are many types of programmed necrosis, it is important to identify those involved in retinal IR to determine the correct treatment. To this end, we used a mouse model of retinal IR and a variety of approaches including RNA-seq data analysis. Our RNA-seq data revealed the rapid development of ischemic pathology in the retina during the first 24 h after reperfusion. We found that at least four types of programmed necrosis including necroptosis, pyroptosis, oxytosis/ferroptosis, and parthanatos are simultaneously involved in retinal IR. Our data suggest that the high activity of the TNF pathway at the early stage of retinal IR leads to early activation of necroptosis while significant activity of other types of programmed necrosis appears later. Our results indicate that TNF, glutamate, and ferrous iron generated by Steap3 may be key players concurrently triggering at least necroptosis, oxytosis/ferroptosis, and parthanatos in ischemic retinal ganglion cells (RGCs). Thus, multiple signaling cascades involved in programmed necrosis should be synchronously targeted for therapeutic purposes to treat retinal IR.
Related collections
Most cited references74
- Record: found
- Abstract: found
- Article: not found
KEGG: kyoto encyclopedia of genes and genomes.
- Record: found
- Abstract: found
- Article: not found
Toward understanding the origin and evolution of cellular organisms

- Record: found
- Abstract: found
- Article: found